Usage#
There are two ways to use NeoFox for annotation of neoantigen candidates with neoantigen features: directly from the command line, docker or programmatically.
Command line#
Neoantigen-Mode#
To call NeoFox from the command line, use the following command. Make sure that the requirements have been added to PATH as described here or add a config file as described below:
neofox --input-file neoantigens_candidates.tsv \
--patient-data patient_data.txt \
--output-folder /path/to/out \
[--output-prefix out_prefix] \
[--organism human|mouse] \
[--rank-mhci-threshold 2.0] \
[--rank-mhcii-threshold 5.0] \
[--num-cpus] \
[--config] \
[--patient-id] \
[--with-all-neoepitopes]
where:
--input-file: tab-separated values table with neoantigen candidates represented by long mutated peptide sequences as described here (extensions .txt and .tsv) or JSON file neoantigens in NeoFox model format as described here (extension .json)--patient-data: a table of tab separated values containing metadata on the patient as described here--output-folder: path to the folder to which the output files should be written--output-prefix: prefix for the output files (optional)--with-all-neoepitopes: output annotations for all MHC-I and MHC-II neoepitopes on all HLA alleles (optional)--rank-mhci-threshold: MHC-I epitopes with a netMHCpan predicted rank greater than or equal than this threshold will be filtered out (optional)--rank-mhcii-threshold: MHC-II epitopes with a netMHCIIpan predicted rank greater than or equal than this threshold will be filtered out (optional)–organism: the organism to which the data corresponds. Possible values: [human, mouse]. Default value: human
--num-cpus: number of CPUs to use (optional)--config: a config file with the paths to dependencies as shown below (optional)--patient-id: patient identifier (optional, this is only relevant if the columnpatientIdentifieris missing in the candidate input file)
PLEASE NOTE THE FOLLOWING HINTS:
If a tumor type has been provided in the patient file, imputated gene expression from TCGA will be annoated and expression-dependent features will be determined with imputated gene expression aswell.
EXAMPLE
This is an example to call NeoFox with a candidate file and obtaining the annotated neoantigen candidates in tabular format:
neofox --input-file neoantigens_candidates.tsv \
--patient-data patient_data.tab \
--output-folder /path/to/out \
--output-prefix test
The optional config file with the paths to the dependencies can look like this:
NEOFOX_REFERENCE_FOLDER=path/to/reference/folder
NEOFOX_RSCRIPT=`which Rscript`
NEOFOX_BLASTP=path/to/blast/bin/blastp
NEOFOX_NETMHCPAN=path/to/netMHCpan/netMHCpan
NEOFOX_NETMHC2PAN=path/to/netMHCIIpan/netMHCIIpan
NEOFOX_MIXMHCPRED=path/to/MixMHCpred/MixMHCpred
NEOFOX_MIXMHC2PRED=path/to/MixMHC2pred/MixMHC2pred_unix
NEOFOX_MAKEBLASTDB=path/to/ncbi-blast/bin/makeblastdb
NEOFOX_PRIME=/path/to/PRIME/PRIME
Neoepitope-Mode#
To call NeoFox over a list neoepitope candidates from the command line, use the following command. The configuration process is similar as described before:
neofox-epitope --input-file neoepitope_candidates.tsv \
--output-folder /path/to/out \
[--patient-data patient_data.txt \]
[--output-prefix out_prefix] \
[--organism human|mouse] \
[--num-cpus] \
[--config] \
where:
--input-file: tab-separated values table with neoepitope candidates represented by mutated peptide sequences as described here (extensions .txt and .tsv)--patient-data: a table of tab separated values containing metadata on the patient as described here--output-folder: path to the folder to which the output files should be written--output-prefix: prefix for the output files (optional)–organism: the organism to which the data corresponds. Possible values: [human, mouse]. Default value: human
--num-cpus: number of CPUs to use (optional)--config: a config file with the paths to dependencies as shown below (optional)
Running from docker#
NOTE: The provided docker recipe is not adapted to Neofox-v1.1.0. Please, use a previous version at the moment if running from docker is required. The docker recipe will be updated soon.
In order to run the command line in a docker image, all of the above applies but some additional steps are required.
If the docker image is named neofox-docker, run as follows: docker run neofox-docker neofox --help
In order to copy the NeoFox input and output data to and from the docker container, a docker volume needs to be created for
mapping a folder in the host to a folder in the container using the -v VOLUME_NAME:ABSOLUTE_FOLDER_IN_CONTAINER argument.
First create a volume:
docker volume create neofox-volume
Identify the folder where the volume is mounted in the host:
$ docker volume inspect neofox-volume
[
{
"CreatedAt": "2021-03-25T21:40:23+01:00",
"Driver": "local",
"Labels": {},
"Mountpoint": "/var/snap/docker/common/var-lib-docker/volumes/neofox-volume/_data",
"Name": "neofox-volume",
"Options": {},
"Scope": "local"
}
]
In the case above the folder is /var/snap/docker/common/var-lib-docker/volumes/neofox-volume/_data.
Copy the input data into that folder.
Now, NeoFox can be run as following by mounting the volume as indicated. Note that the output folder needs to be specified within the volume, if the output from NeoFox should be recovered.
docker run -v neofox-volume:/app/data neofox-docker \
neofox --input-file /app/data/test_model_file.txt \
--patient-data /app/data/test_patient_info.txt \
--output-folder /app/data/output
API#
NeoFox can be used programmatically and by that integrated into existing tools. Here, we will explain the use of NeoFox by API in short with the help of a dummy example that includes building models from scratch (For a detailed description, please refer to this notebook). The models can be created based on files too. In this case, ignore step 2-5 and refer to the note on the bottom of this paragraph.
NeoFox can be executed as follows.
annotations = NeoFox(neoantigens=[neoantigen], patients=[patient], num_cpus=2).get_annotations()
Follow the next detailed steps.
NOTE: the environment variables need to be loaded. Hint: use dotenv.load_dotenv().
Create a neoantigen object#
Create a neoantigen candidate using NeoantigenFactory.
The data will be internally validated.
Additional annotations with custom names are supported.
from neofox.model.factories import NeoantigenFactory
# create a neoantigen candidate using the factory
neoantigen = NeoantigenFactory.build_neoantigen(
mutated_xmer="AAAAAAAAAAAAARAAAAAAAAAAAAA",
wild_type_xmer="AAAAAAAAAAAAAMAAAAAAAAAAAAA",
patient_identifier="Ptx",
rna_expression=0.52,
rna_variant_allele_frequency=0.88,
dna_variant_allele_frequency=0.29,
my_custom_annotation="add any custom annotation as additional fields with any name"
)
Create a patient object#
In order to parse MHC alleles and being able to normalize them into a standard nomenclature, load the following resources.
from neofox.references.references import ReferenceFolder
reference_folder = ReferenceFolder(organism='human')
To parse mouse H-2 alleles use ReferenceFolder(organism='mouse').
Create a patient model based on models for MHC I and MHC II alleles.
from neofox.model.factories import PatientFactory
patient = PatientFactory.build_patient(
identifier="Ptx",
mhc_alleles=["HLA-A*01:01:02:03N", "HLA-A*01:02:02:03N", "HLA-B*01:01:02:03N", "HLA-B*01:01:02:04N", "HLA-C*01:01"],
mhc2_alleles=["HLA-DPA1*01:01", "HLA-DPA1*01:02", "HLA-DPB1*01:01", "HLA-DPB1*01:01", "HLA-DRB1*01:01", "HLA-DRB1*01:01"],
mhc_database=reference_folder.get_mhc_database()
)
**WARNING**\ : alleles in homozygous state need to be provided twice, otherwise they are considered as hemizygous.
For instance ``["HLA-A*01:01"]`` would be interpreted as hemizygous and ``["HLA-A*01:01", "HLA-A*01:01"]`` as homozygous.
Create a neoepitope object#
Create a neoepitope candidate as indicated below. The data will be internally validated. Additional annotations with custom names are supported.
from neofox.model.factories import NeoepitopeFactory
from neofox.references.references import ReferenceFolder
hla_database = ReferenceFolder(organism='human').get_mhc_database()
# create a neoantigen candidate using the factory
neoepitope = NeoepitopeFactory.build_neoepitope(
mutated_peptide="AAAARAAAA",
wild_type_peptide="AAAAMAAAA",
allele_mhc_i="HLA-A*01:01",
rna_expression=0.52,
rna_variant_allele_frequency=0.88,
dna_variant_allele_frequency=0.29,
my_custom_annotation="add any custom annotation as additional fields with any name",
organism='human',
mhc_database=hla_database
)
Run NeoFox#
Run NeoFox by passing the neoantigen and patients object to get the neoantigen features.
The output is a list of type NeoantigenAnnotations:
from neofox.neofox import NeoFox
annotated_neoantigens = NeoFox(neoantigens=[neoantigen], patients=[patient], num_cpus=2).get_annotations()
where:
- `neoantigens`: a list of neoantigen objects
- `patients`: a list of patient objects
- `num_cpus`: number of CPUs to use (*optional*)
HINT: process multiple neoantigens by passing a list of neoantigens and a list of patients to NeoFox().get_annotations().
Run NeoFox for neoepitopes#
Run NeoFox by passing the neoepitope and patients object to get the neoantigen features.
The output is a list of type PredictedEpitope:
from neofox.neofox_epitope import NeoFoxEpitope
annotated_neoepitopes = NeoFoxEpitope(neoepitopes=[neoepitope], patients=[patient], num_cpus=2).get_annotations()
where:
- `neoepitopes`: a list of neoepitope objects
- `patients`: a list of patient objects
- `num_cpus`: number of CPUs to use (*optional*)
HINT: process multiple neoepitopes by passing a list of neoepitopes and a list of patients to NeoFoxEpitope().get_annotations().
Data transformation#
Depending on the use case, the user can transform the resulting neoantigen feature annotations into a Pandas data frame or into JSON format, as described here.
from neofox.model.conversion import ModelConverter
# Pandas data frame
annotations_table = ModelConverter.annotations2neoantigens_table(neoantigens=annotated_neoantigens)
# JSON
neoantigen_json = ModelConverter.objects2json(model_objects=annotated_neoantigens)
The same is applicable for a list of input patients.
# Pandas data frame
patients_table = ModelConverter.patients2table(patients=patients)
# JSON
patients_json = ModelConverter.objects2json(model_objects=patients)
instead of creating neoantigen or patient models, tabular or json files containing this information can be passed:
The neoantigen candidates can be provided in candidate file format
model_file = "/path/to/neoantigen_candidates.tab"
neoantigens = ModelConverter.parse_neoantigens_file(neoantigens_file=model_file)
or in JSON format.
json_file = "/path/to/neoantigen_candidates.json"
neoantigens = ModelConverter.parse_neoantigens_json_file(json_file=json_file)
The patient information should be provided in tabular format
patient_file = "/path/to/patients.tab"
patients = ModelConverter.parse_patients_file(patients_data)
Then, run NeoFox as explained before by calling:
neoantigens_annotated = NeoFox(neoantigens=neoantigens, patients=patients, num_cpus=2).get_annotations()
Performance#
As indicated above NeoFox can run in parallel using the parameter --num-cpus.
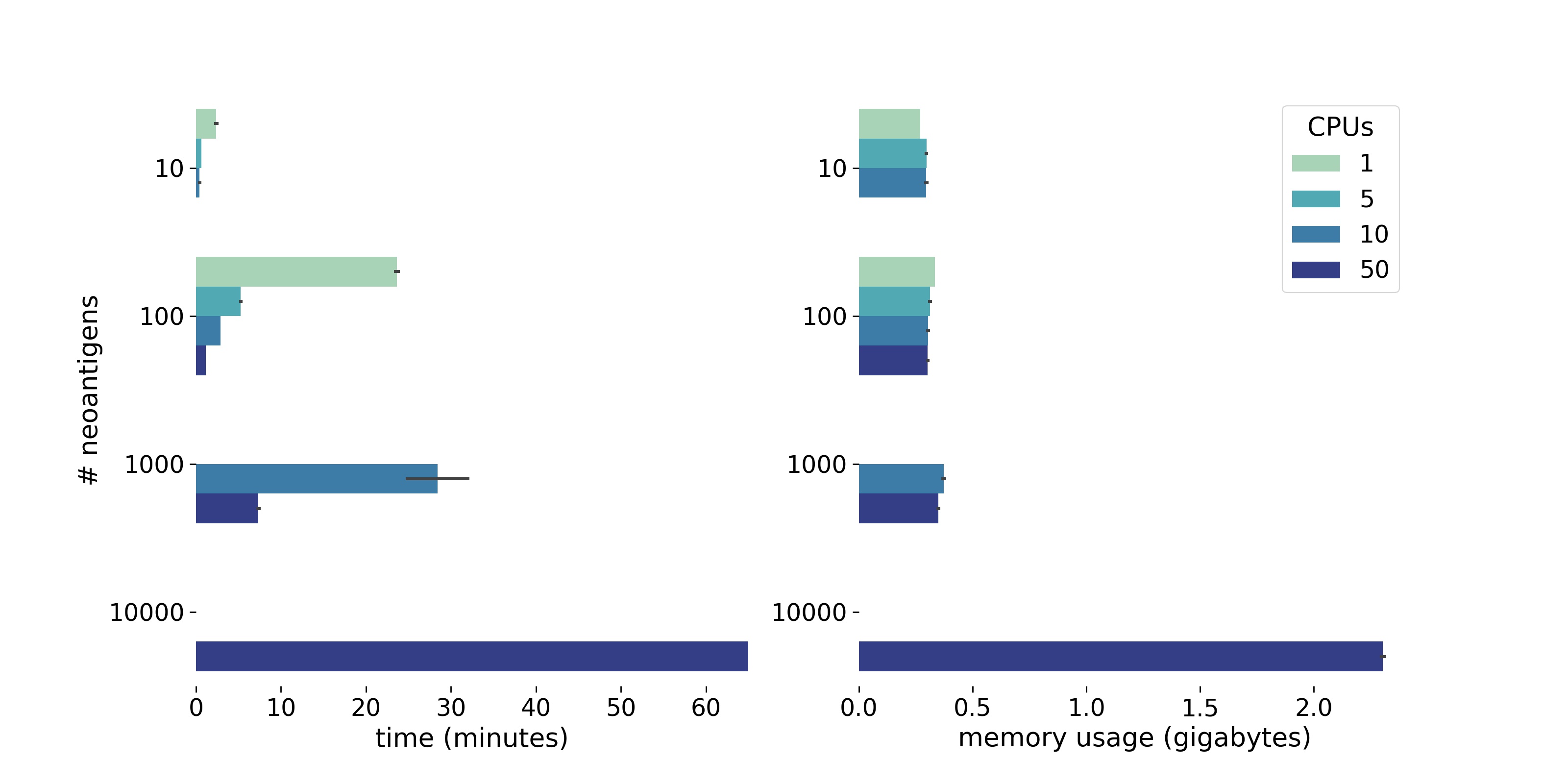
Each CPU will process one neoantigen candidate at a time, thus NeoFox uses only as many CPUs as candidats are to be processed.
We processed several simulated datasets with 10, 100, 1000 and 10000 neoantigen candidates on 1, 5, 10 and 50 CPUs. We obtained that the average time to process a single candidate in a single CPU takes 37.516 seconds, with a standard deviation of 6.739 seconds. No significant overhead due to parallelization was observed. In terms of memory the application uses less than 0.5 GB for up to 1000 neoantigen candidates irrespective of the number of CPUs used. The memory use grows to around 2.5 GB when processing 10000 candidates.
If either MHC I or II alleles are not provided at all for a given patient the computation will be lighter as no annotations run for the missing MHC. Likewise, if the optional tools are unset performance improves.