Data models#
Protocol Buffers is employed to model Neofox’s input and output data: neoantigens, Major Histocompatibility Complex (MHC) alleles, patients and output annotations.
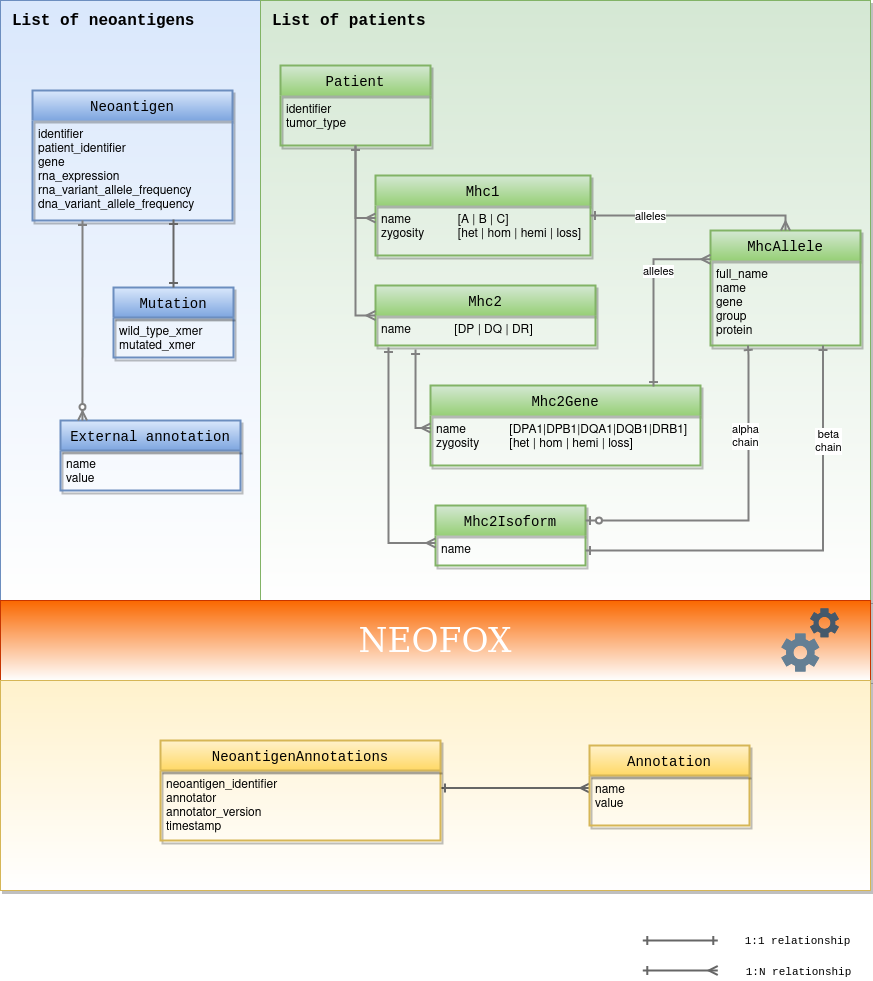
Table of Contents#
neoantigen.proto#
Annotation#
This is a generic class to hold annotations from Neofox
Field |
Type |
Label |
Description |
|---|---|---|---|
name |
The name of the annotation |
||
value |
The value of the annotation |
Annotations#
A set of annotations for a neoantigen candidate
Field |
Type |
Label |
Description |
|---|---|---|---|
annotations |
repeated |
List of annotations |
|
annotator |
The annotator |
||
annotatorVersion |
The version of the annotator |
||
timestamp |
A timestamp determined when the annotation was created |
||
resources |
repeated |
List of resources |
Mhc1#
Models MHC I alleles related to the same MHC I gene, i.e. 2 alleles/2 isoforms per gene
Field |
Type |
Label |
Description |
|---|---|---|---|
name |
MHC I gene name |
||
zygosity |
Zygosity of the gene |
||
alleles |
repeated |
The alleles of the gene (0, 1 or 2) |
Mhc2#
Models MHC II alleles related to the same MHC II protein, i.e. 4 isoforms related to 2 genes with 2 alleles each
Field |
Type |
Label |
Description |
|---|---|---|---|
name |
MHC II molecule name |
||
genes |
repeated |
List of MHC II genes |
|
isoforms |
repeated |
Different combinations of MHC II alleles building different isoforms |
Mhc2Gene#
MHC II gene
Field |
Type |
Label |
Description |
|---|---|---|---|
name |
MHC II gene name |
||
zygosity |
Zygosity of the gene |
||
alleles |
repeated |
The alleles of the gene (0, 1 or 2) |
Mhc2Isoform#
MHC II isoform
Field |
Type |
Label |
Description |
|---|---|---|---|
name |
Name to refer to the MHC II isoform |
||
alphaChain |
The alpha chain of the isoform |
||
betaChain |
The beta chain of the isoform |
MhcAllele#
MHC allele representation. It does not include non synonymous changes to the sequence, changes in the non coding region or changes in expression. See http://hla.alleles.org/nomenclature/naming.html for details
Field |
Type |
Label |
Description |
|---|---|---|---|
fullName |
HLA full name as provided by the user (e.g.: HLA-DRB1*13:01:02:03N). This will be parsed into name, gene and group. Any digit format is allowed for this field (ie: 4, 6 or 8 digits), 2 digits names are not specific enough for our purpose and thus invalid |
||
name |
A specific HLA protein (e.g. HLA-DRB113:01). Alleles whose numbers differ in group and protein must differ in one or more nucleotide substitutions that change the amino acid sequence of the encoded protein. This name is normalized to avoid different representations of the same allele. For instance both HLA-DRB113:01 and HLA-DRB113:01:02:03N will be transformed into their normalised version HLA-DRB1*13:01. This name is also truncated to 4 digits. 2 digits names are not specific enough for our purpose and thus invalid |
||
gene |
The gene from either MHC I or II (e.g. DRB1, A) (this information is redundant with the Mhc1Gene.name and Mhc2Gene.name but it is convenient to have this at this level too, code will check for data coherence) |
||
group |
A group of alleles defined by a common serotype ie: Serological antigen carried by an allotype (e.g. 13 from HLA-DRB1*13) |
||
protein |
A specific protein (e.g.: 02 from HLA-DRB1*13:02) |
Neoantigen#
A neoantigen minimal definition
Field |
Type |
Label |
Description |
|---|---|---|---|
patientIdentifier |
Patient identifier |
||
gene |
The HGNC gene symbol or gene identifier |
||
position |
repeated |
The amino acid position within the neoantigen candidate sequence. 1-based, starting in the N-terminus |
|
wildTypeXmer |
Amino acid sequence of the WT corresponding to the neoantigen candidate sequence (IUPAC 1 letter codes) |
||
mutatedXmer |
Amino acid sequence of the neoantigen candidate (IUPAC 1 letter codes) |
||
rnaExpression |
Expression value of the transcript from RNA data. Range [0, +inf]. |
||
imputedGeneExpression |
Expression value of the transcript from TCGA data. Range [0, +inf]. |
||
dnaVariantAlleleFrequency |
Variant allele frequency from the DNA. Range [0.0, 1.0] |
||
rnaVariantAlleleFrequency |
Variant allele frequency from the RNA. Range [0.0, 1.0] |
||
neofoxAnnotations |
The NeoFox neoantigen annotations |
||
externalAnnotations |
repeated |
List of external annotations |
|
neoepitopesMhcI |
repeated |
List of predicted neoepitopes for MHC-I with feature annotation (optional) |
|
neoepitopesMhcII |
repeated |
List of predicted neoepitopes for MHC-II with feature annotation (optional) |
Patient#
The metadata required for analysis for a given patient + its patient identifier
Field |
Type |
Label |
Description |
|---|---|---|---|
identifier |
Patient identifier |
||
tumorType |
Tumor entity in TCGA study abbrevation style as described here: https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations |
||
mhc1 |
repeated |
MHC I classic molecules |
|
mhc2 |
repeated |
MHC II classic molecules |
PredictedEpitope#
Field |
Type |
Label |
Description |
|---|---|---|---|
position |
Not sure that we need this… this is in the old PredictedEpitope model |
||
mutatedPeptide |
The mutated peptide |
||
wildTypePeptide |
Closest wild type peptide |
||
alleleMhcI |
MHC I allele |
||
isoformMhcII |
MHC II isoform |
||
core |
MHCII core part of the peptide ligand that primarily interacts with the MHC binding groove, predicted by NetMHCpan/NetMHCIIpan |
||
affinityMutated |
MHC binding affinity for the mutated peptide. This value is estimated with NetMHCpan in case of MHC-I peptides and NetMHCIIpan in cas of MHC-II peptides |
||
rankMutated |
MHC binding rank for the mutated peptide. This value is estimated with NetMHCpan in case of MHC-I peptides and NetMHCIIpan in cas of MHC-II peptides |
||
affinityWildType |
MHC binding affinity for the wild type peptide. This value is estimated with NetMHCpan in case of MHC-I peptides and NetMHCIIpan in cas of MHC-II peptides |
||
rankWildType |
MHC binding rank for the wild type peptide. This value is estimated with NetMHCpan in case of MHC-I peptides and NetMHCIIpan in cas of MHC-II peptides |
||
neofoxAnnotations |
The NeoFox neoantigen annotations |
||
patientIdentifier |
Patient identifier |
||
gene |
The HGNC gene symbol or gene identifier |
||
rnaExpression |
Expression value of the transcript from RNA data. Range [0, +inf]. |
||
imputedGeneExpression |
Expression value of the transcript from TCGA data. Range [0, +inf]. |
||
dnaVariantAlleleFrequency |
Variant allele frequency from the DNA. Range [0.0, 1.0] |
||
rnaVariantAlleleFrequency |
Variant allele frequency from the RNA. Range [0.0, 1.0] |
||
externalAnnotations |
repeated |
External annotations for neoepitope mode. |
Resource#
This is a class to track the version of an annotation resource
Field |
Type |
Label |
Description |
|---|---|---|---|
name |
The name of the resource |
||
version |
The version of the resource |
||
url |
The URL of the resource if applicable |
||
hash |
The MD5 hash of the resource if applicable. This may be used when version is not available |
||
download_timestamp |
The timestamp when the download happened |
Mhc1Name#
Valid names for MHC I classic genes Mus musculus gene names are preceded by the prefix H2 to avoid naming collisions.
Mhc2GeneName#
Valid names for MHC II classic genes. DRA is not included in this list as it does not have much variability in the population and for our purpose is considered constant. For Mus musculus we do not represent alpha and beta chains as they are homozygotes at all their MHC loci. Hence, they can be treated as a single gene, like DR is for HLA. See http://www.imgt.org/IMGTrepertoireMH/Polymorphism/haplotypes/mouse/MHC/Mu_haplotypes.html Mus musculus gene names are preceded by the prefix H2 to avoid naming collisions.
Mhc2Name#
Valid names for MHC II classic molecules
Zygosity#
The zygosity of a given gene
Name |
Number |
Description |
|---|---|---|
HOMOZYGOUS |
0 |
Two equal copies of the gene |
HETEROZYGOUS |
1 |
Two different copies of the gene |
HEMIZYGOUS |
2 |
Only one copy of the gene |
LOSS |
3 |
No copy of the gene |
Scalar Value Types#
.proto Type |
Notes |
C++ |
Java |
Python |
Go |
C# |
PHP |
Ruby |
|---|---|---|---|---|---|---|---|---|
double |
double |
float |
float64 |
double |
float |
Float |
||
float |
float |
float |
float32 |
float |
float |
Float |
||
Uses variable-length encoding. Inefficient for encoding negative numbers – if your field is likely to have negative values, use sint32 instead. |
int32 |
int |
int |
int32 |
int |
integer |
Bignum or Fixnum (as required) |
|
Uses variable-length encoding. Inefficient for encoding negative numbers – if your field is likely to have negative values, use sint64 instead. |
int64 |
long |
int/long |
int64 |
long |
integer/string |
Bignum |
|
Uses variable-length encoding. |
uint32 |
int |
int/long |
uint32 |
uint |
integer |
Bignum or Fixnum (as required) |
|
Uses variable-length encoding. |
uint64 |
long |
int/long |
uint64 |
ulong |
integer/string |
Bignum or Fixnum (as required) |
|
Uses variable-length encoding. Signed int value. These more efficiently encode negative numbers than regular int32s. |
int32 |
int |
int |
int32 |
int |
integer |
Bignum or Fixnum (as required) |
|
Uses variable-length encoding. Signed int value. These more efficiently encode negative numbers than regular int64s. |
int64 |
long |
int/long |
int64 |
long |
integer/string |
Bignum |
|
Always four bytes. More efficient than uint32 if values are often greater than 2^28. |
uint32 |
int |
int |
uint32 |
uint |
integer |
Bignum or Fixnum (as required) |
|
Always eight bytes. More efficient than uint64 if values are often greater than 2^56. |
uint64 |
long |
int/long |
uint64 |
ulong |
integer/string |
Bignum |
|
Always four bytes. |
int32 |
int |
int |
int32 |
int |
integer |
Bignum or Fixnum (as required) |
|
Always eight bytes. |
int64 |
long |
int/long |
int64 |
long |
integer/string |
Bignum |
|
bool |
boolean |
boolean |
bool |
bool |
boolean |
TrueClass/FalseClass |
||
A string must always contain UTF-8 encoded or 7-bit ASCII text. |
string |
String |
str/unicode |
string |
string |
string |
String (UTF-8) |
|
May contain any arbitrary sequence of bytes. |
string |
ByteString |
str |
[]byte |
ByteString |
string |
String (ASCII-8BIT) |